Quantification of Human eRNA Atlas
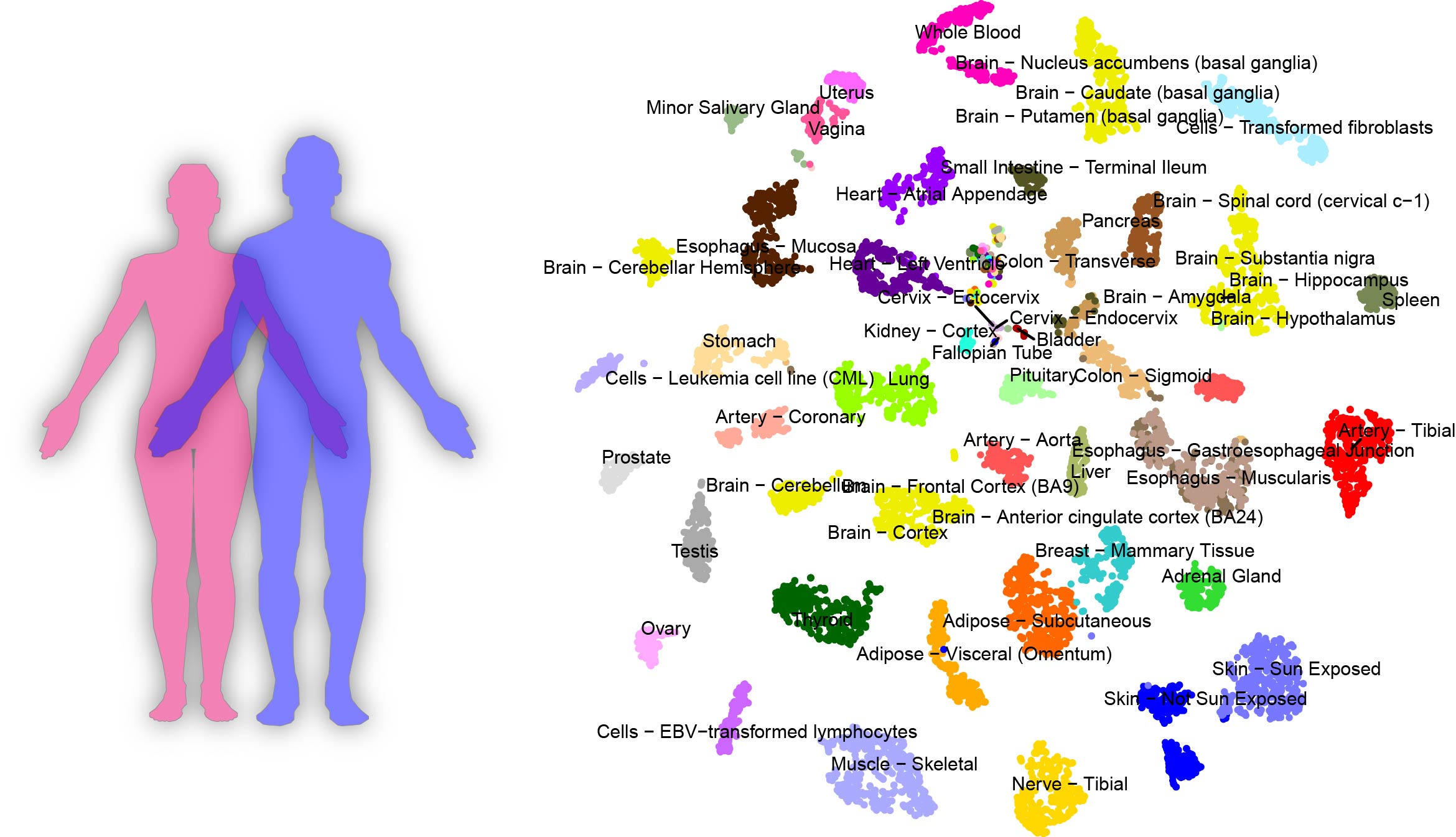
HeRA identified 45,411 eRNA from 9,577 samples across 54 human tissues from the Genotype-Tissue Expression (GTEx) Project (hg19). Annotations of enhancers were collected from the Encyclopedia of DNA Elements Project (ENCODE), Functional Annotation of the Mammalian Genome (FANTOM) Project and Roadmap Epigenomics Project. We defined ±3kb around the middle loci of the enhancer as the eRNA region. We filtered out eRNA regions that overlapped with known annotations (e.g., CDS, non-coding RNAs) or which were close to annotated regions (<1kb). We mapped these RNA-seq reads to eRNA regions and used reads pre million (RPM) to normalize eRNA expressions. eRNAs have mean expression value (RPM ≥ 1) in at least one tissue were defined as detectable eRNAs.
HeRA provides four functional modules
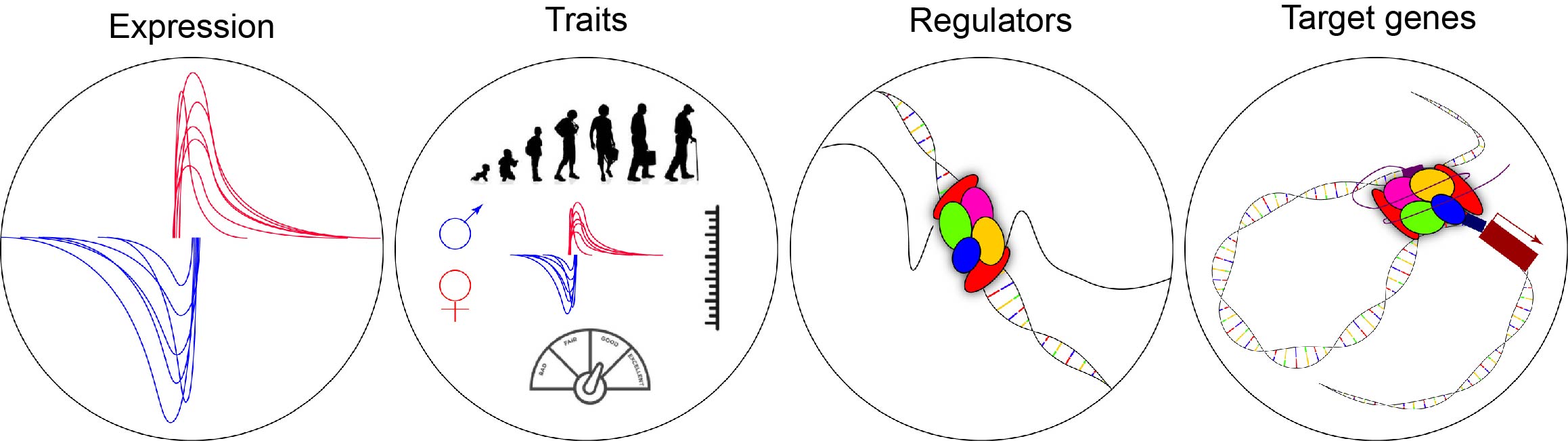
HeRA provides four functional modules to display eRNA expression, trait-related eRNAs, putative regulators and target genes of eRNAs.
The expression module enables users to query the eRNA expression landscape for each sample across tissues.
The traits module enables users to query trait-associated eRNAs, including those associated with age, gender, and body mass index (BMI).
The regulators module enables users to query transcription factors that are significantly associated with eRNAs in each tissue.
The target genes module enables users to query target genes (within 1 MB) that may be regulated by eRNAs in each tissue.
HeRA provides multiple options for eRNA querying.

HeRA supplies multiple options for eRNA querying.
(A) All four modules provide tissue-select options through which users can select one or multiple tissues (default = 54).
(B) All four modules provide three options for eRNA querying: eRNA ID, genomic region, and gene symbol.
NOTE: in Expression and Traits module, a gene symbol is used to query eRNAs within 1MB of the transcription start site of a given gene;
in Regulations module, only transcription factors are allowed to query for a gene symbol;
in Target genes module, a gene symbol will query eRNAs within 1MB distance of the transcription start site.
(C) In the Traits module, users can select one or multiple traits.